Dear All (and with thanks to Lance Price for co-authoring this deeply wonkish newsletter),
Today we turn to the idea of examining livestock manure for the presence of antibiotic resistance genes. You can easily see how this idea plays into the question of how to track and control AMR.
Before we dive into the paper, however, a few definitions are needed. Here we go!
- Genome: All of the DNA of a specific organism.
- It also refers to the idea of all the variants of the genome of multiple members of a given species.
- This is a familiar idea … we use it all the time.
- Metagenome: All of the DNA found in material from a mixture of species.
- Imagine taking a sample of dirt (or manure, as we’ll do a bit later on), and sequencing everything you find.
- Yes, it’s a jumble. But, with the knowledge of all the genomes available to us and with the help of a supercomputer (or two), you can make good guesses about what goes with what and then you can create the next item on our list…
- Metagenome-assembled genomes (MAGs): Here, the supercomputer has done its best to bring together all the observed bits of sequence data (these are called contigs) and lay them out in ways that appear to match the typical genome of known organisms such as E. coli.
So with those ideas available to us, let’s now look at the paper that triggered this newsletter. Here are the links you need:
- (the primary paper) Li B, Jiang L, Johnson T, Wang G, Sun W, Wei G, et al. Global health risks lurking in livestock resistome. Science Advances. 2025;11(26); https://doi.org/10.1126/sciadv.adt8073.
- (three papers offering ideas about ways to rank possible antibiotic resistance genes, aka ARGs)
- Martínez JL, Coque TM, Baquero F. What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol. 2015;13(2):116–23; https://doi.org/10.1038/nrmicro3399.
- Zhang A-N, Gaston JM, Dai CL, Zhao S, Poyet M, Groussin M, et al. An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nature Communications. 2021;12(1):4765; https://doi.org/10.1038/s41467-021-25096-3.
- Zhang Z, Zhang Q, Wang T, Xu N, Lu T, Hong W, et al. Assessment of global health risk of antibiotic resistance genes. Nat Commun. 2022;13(1):1553; https://doi.org/10.1038/s41467-022-29283-8.
- (ARGs-OAP = ARGs Online Analysis Pipeline, a ranking of ARGs) Yin X, Zheng X, Li L, Zhang A-N, Jiang X-T, Zhang T. ARGs-OAP v3.0: Antibiotic-Resistance Gene Database Curation and Analysis Pipeline Optimization. Engineering. 2023;27:234–41; https://doi.org/10.1016/j.eng.2022.10.011.
—
The core idea of the primary paper turns out to be simple: the authors “analyzed 4,017 livestock manure metagenomes from 26 countries, to illuminate the global patterns of livestock resistomes and the prevalence and distribution of clinically critical ARG subtypes.” And they did indeed pull together a huge survey by mining available databases … those 4,017 metagenomes included 1,115 chicken, 1,102 bovine, and 1,759 swine metagenomes. Here’s the map:
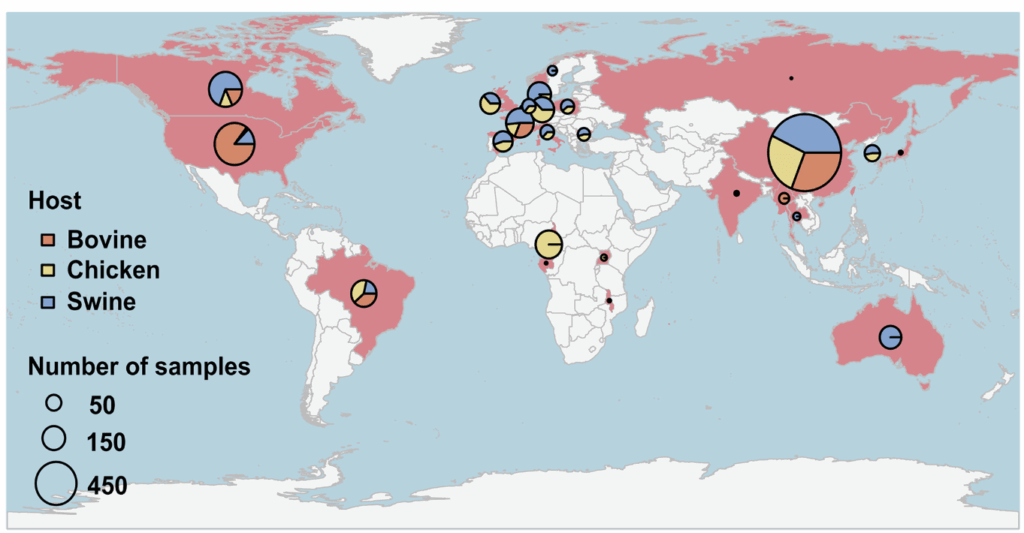
With these data in hand, the authors assembled 123,872 MAGs and found that 12,069 of them contained 563 different ARGs. This step involves some deep thinking about the meaning of “antibiotic resistance gene” and this is where Martinez 2015, Zhang 2021, Zhang 2022, and Yin 2023 come into play.
As noted by those background papers, previous metagenomic studies have shown that animal wastes, and even pristine environments like untouched soils, harbor a vast array of ARGs. However, most of these genes are likely embedded in the chromosomes of environmental microbes that pose little or no threat to human health. Critically, ARGs are only relevant to the extent that they are: (i) present in pathogens, (ii) actually able to cause resistance to one or more relevant drugs, and/or (iii) mobile across/between organisms so that the mechanism of resistance can spread.
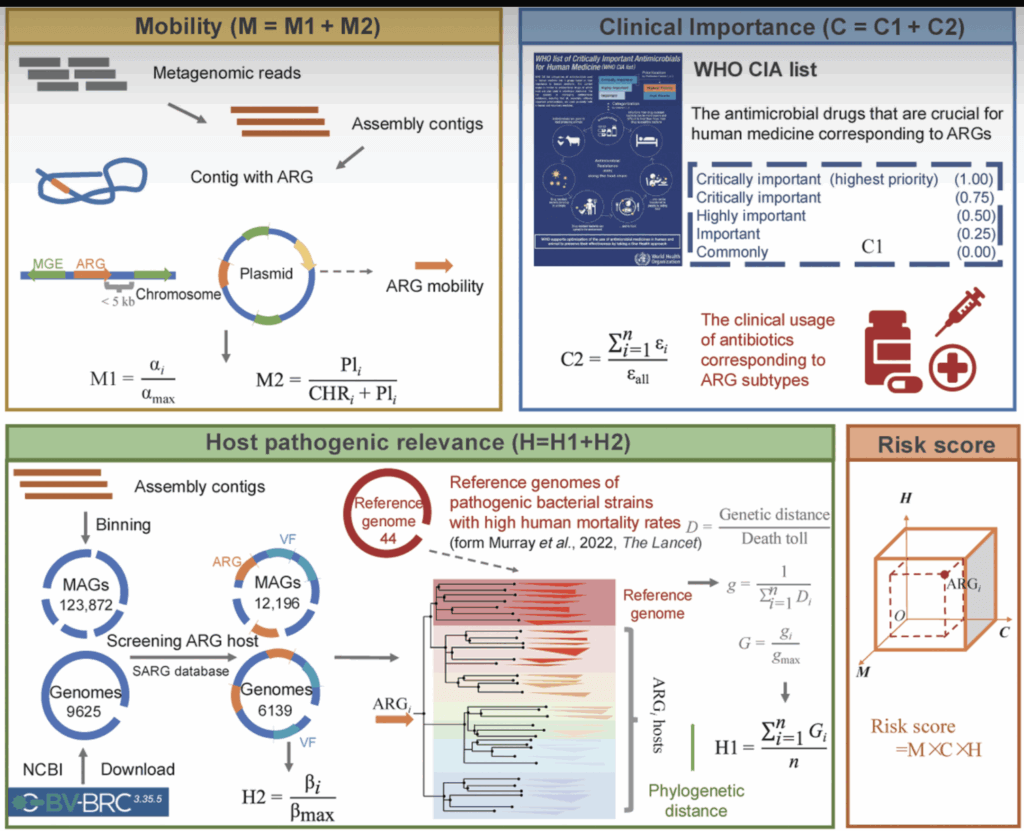
Using this conceptual framework, Li et al. have integrated ideas from the background papers. Using bioinformatic tools, they develop data on the abundance of ARGs in a given metagenome and the diversity of the ARGs. From this, they compute an integrated numeric view of risk via a score that ranges from 0 to 4 (see figure below our signatures for the math behind the scoring system).
—
So, what did they learn? This is the key figure and it requires a bit of study:
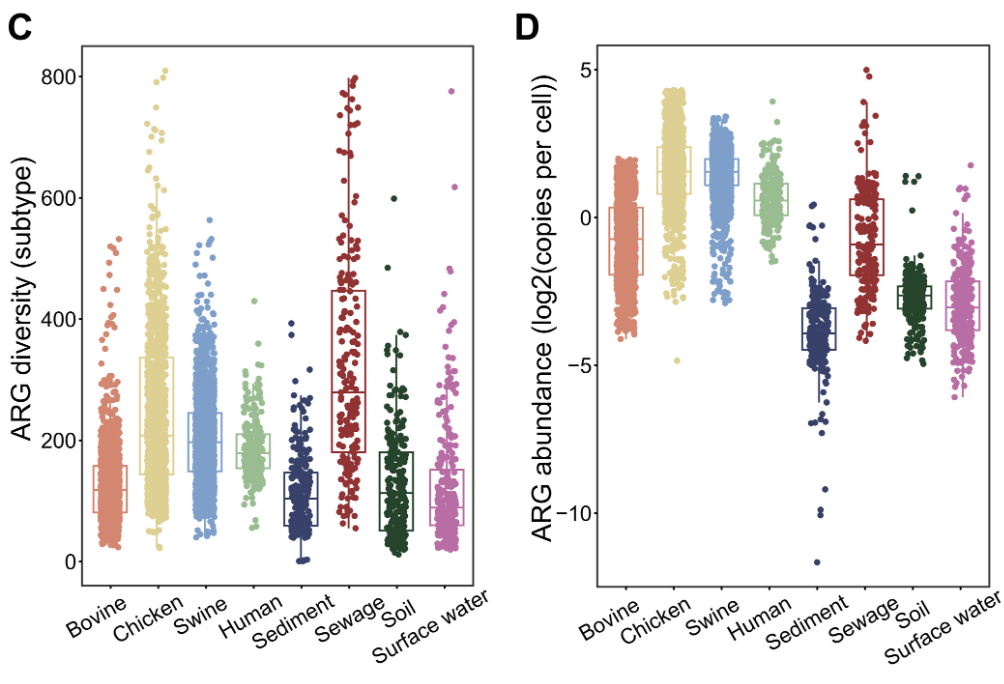
The left-hand panel is ARG diversity and the right is ARG abundance. Note also the new comparator groups on the x-axes: the authors have included data from 1,117 metagenomes from human feces, sewage, sediment, soil, and surface water.
Now, note the small boxes drawn in each column: these appear to represent median and inter-quartile ranges. Also note that the y-axis in the right-hand (abundance) figure is logarithmic … small shifts mean a lot.
With these features noted, you’ll see that chickens and swine easily lead the livestock pack in terms of both diversity and abundance. Overall, there is a strong hierarchy when you consider both diversity and abundance of ARGs in livestock: with a ranking of chicken > pig >> cattle.
—
But what about the nature of the ARGs themselves? Well, it turns out that livestock and humans share similar resistome patterns; soil, sediment, and water share a different set of similar resistome patterns; and sewage spans the gap. This is seen in this principal component analysis diagram (PCoA: in brief, all the variation in pair of data sets is collapsed into a 2-dimensional graph):
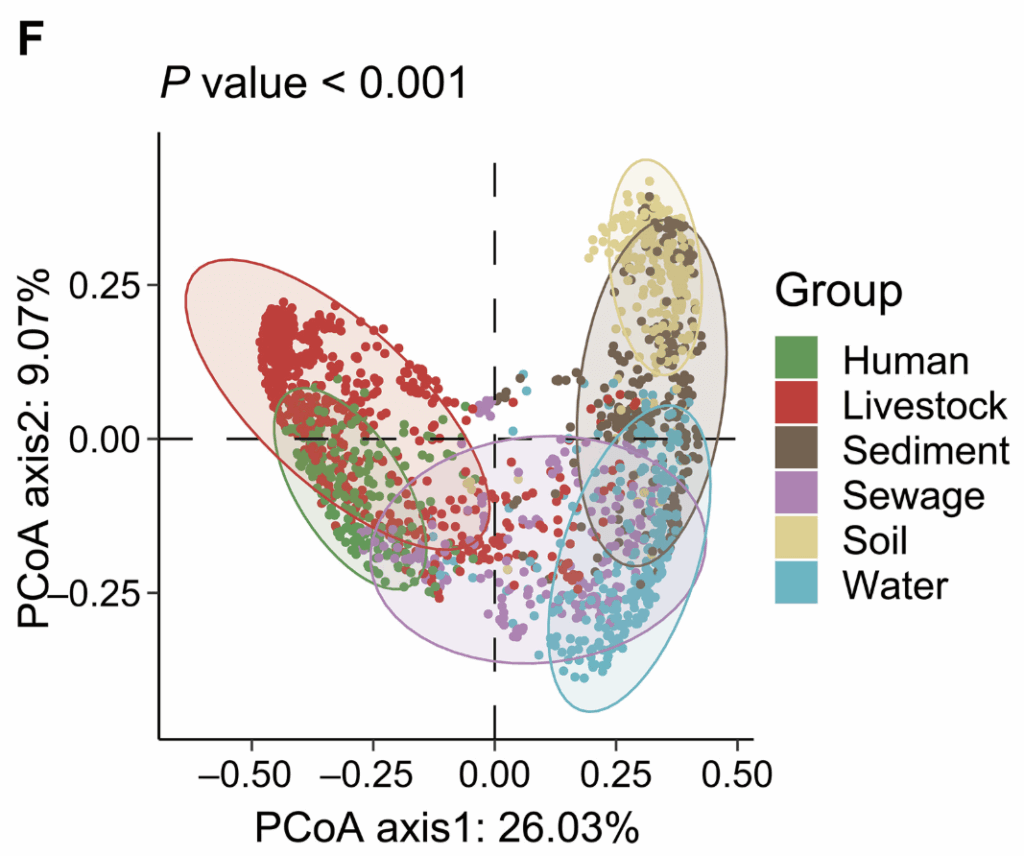
And when you think about it, you can easily see why it is logical that these groupings should be true.
—
Finally, the authors used all the data to construct global heat maps showing the relative risk posed by the ARGs that they observed:
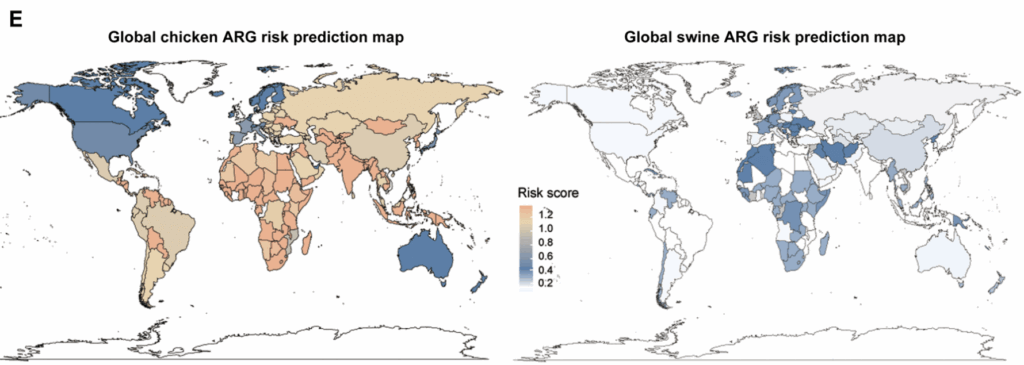
The scale here is the 0-4 risk score mentioned above. You can see immediately that the scores are overall higher for chickens than swine (there’s no red anywhere on the swine figure at right) with the highest scores for chickens occurring in South America, Africa, and Asia. The range in the figure for swine is a bit more compressed numerically but you can see the highest scores are in Africa and Western Europe.
—
Finally, what does this all mean? What next steps are most relevant?
As noted above, this paper is a significant advance over previous metagenomic studies by providing quantitative estimates for animal production around the world. However, it is limited by the dearth of actual data from some of the poorest regions of the world. For example, the study identified Africa as one of the highest-risk regions for antimicrobial resistance in livestock, yet the geography figure above reveals a striking lack of sampling across the continent. Most of the African data come from a single country – Cameroon – and primarily from poultry.
It is also important to note that the authors relied on a machine-learning model to estimate resistome risk elsewhere. The model is logical in that it incorporates factors such as antimicrobial use in food animal production but this limitation of the study is, in our view, a clear call to action. In the absence of empirical antimicrobial use data, the model draws on inputs from previous studies that themselves rely on modeled estimates. This practice of feeding models with modeled data introduces layers of uncertainty that should not be overlooked. To improve future risk assessments, we urgently need more direct measurements of antimicrobial use and microbial sampling across Africa and other under-resourced regions.
With these limitations in mind, this study reinforces the concern that food animals can serve as reservoirs of high-risk ARGs. This is particularly true in low- and middle-income countries where widespread antimicrobial use, close human–animal contact, and inadequate water, sanitation, and hygiene conditions may amplify public health risks. We commend the authors for taking a strong step forward in applying metagenomics to a One Health context and we hope that these results will be useful in creating awareness and action!
All best wishes, John & Lance
John H. Rex, MD | Chief Medical Officer, F2G Ltd. | Operating Partner, Advent Life Sciences. Follow me on Twitter: @JohnRex_NewAbx. See past newsletters and subscribe for the future: https://amr.solutions/blog/. All opinions are my own.
Lance B. Price, PhD | Co-director, Antibiotic Resistance Action Center | Professor, Milken Institute School of Public Health, George Washington University. E: lprice@gwu.edu; Twitter @battlesuperbugs. All opinions are my own.
—
Figure from Li 2025 showing how Mobility, Clinical Importance, and Host (pathogen) are combined to produce a Risk Score from 0 to 4:
- The AMR Industry Alliance is again offering a Stewardship Prize of 10,000 CHF to recognize innovative approaches to combating AMR in low-to-moderate-income countries. This year’s prize focuses on utilizing diagnostics in stewardship efforts. Applications are due by 1 Sep 2025. Go here for details and to apply.
- NNF (Novo Nordisk Foundation) have announced their “Challenge Programme 2026 – Unravelling the Pathways of Human Invasive Fungal Diseases. The call seeks applications from EU-centered consortia (global partners are possible) for research in 4 areas: (i) fungal virulence factors, (ii) host-pathogen interactions, (iii) mechanisms of anti-fungal resistance, and (iv) fungal disease markers. Applications are due by 8 Oct 2025. Go here for details.
- ENABLE-2 has continuously open calls for both its Hit-to-Lead program as well as its Hit Identification/Validation incubator. Applicants must be academics and non-profits in Europe due to restrictions from the funders. Applications are evaluated in cycles … see the website for details on current timing for reviews.
- CARB-X will have two calls during 2025 that span two areas: (i) Small molecules for Gram-negatives (the focus is on Pseudomonas aeruginosa) and (ii) Diagnostics for typhoid (the focus is diagnosis of acute infections in 60 minutes or less). See this 26 Feb 2025 newsletter for a discussion of the call and go here for the CARB-X webpage on the call. The first cycle is now closed (it ran16-30 April 2025); the 2nd round will be open 1-12 Dec 2025.
- BARDA’s long-running BAA (Broad Agency Announcement) for medical countermeasures (MCMs) for chemical, biological, radiological, and nuclear (CBRN) threats, pandemic influenza, and emerging infectious diseases is now BAA-23-100-SOL-00004 and offers support for both antibacterial and antifungal agents (as well as antivirals, antitoxins, diagnostics, and more). Note especially these Areas of Interest: Area 3.1 (MDR Bacteria and Biothreat Pathogens), Area 3.2 (MDR Fungal Infections), and Area 7.2 (Antibiotic Resistance Diagnostics for Priority Bacterial Pathogens). Although prior BAAs used a rolling cycle of 4 deadlines/year, the updated BAA released 26 Sep 2023 has a 5-year application period that ends 25 Sep 2028 and is open to applicants regardless of location: BARDA seeks the best science from anywhere in the world! See also this newsletter for further comments on the BAA and its areas of interest.
- HERA Invest was launched August 2023 with €100 million to support innovative EU-based SMEs in the early and late phases of clinical trials. Part of the InvestEU program supporting sustainable investment, innovation, and job creation in Europe, HERA Invest is open for application to companies developing medical countermeasures that address one of the following cross-border health threats: (i) Pathogens with pandemic or epidemic potential, (ii) Chemical, biological, radiological and nuclear (CBRN) threats originating from accidental or deliberate release, and (iii) Antimicrobial resistance (AMR). Non-dilutive venture loans covering up to 50% of investment costs are available. A closing date is not posted insofar as I can see — applications are accepted on a rolling basis; go here for more details.
- The AMR Action Fund is open on an ongoing basis to proposals for funding of Phase 2 / Phase 3 antibacterial therapeutics. Per its charter, the fund prioritizes investment in treatments that address a pathogen prioritized by the WHO, the CDC and/or other public health entities that: (i) are novel (e.g., absence of known cross-resistance, novel targets, new chemical classes, or new mechanisms of action); and/or (ii) have significant differentiated clinical utility (e.g., differentiated innovation that provides clinical value versus standard of care to prescribers and patients, such as safety/tolerability, oral formulation, different spectrum of activity); and (iii) reduce patient mortality. It is also expected that such agents would have the potential to strongly address the likely requirements for delinked Pull incentives such as the UK (NHS England) subscription pilot and the PASTEUR Act in the US. Submit queries to contact@amractionfund.com.
- INCATE (Incubator for Antibacterial Therapies in Europe) is an early-stage funding vehicle supporting innovation vs. drug-resistant bacterial infections. The fund provides advice, community, and non-dilutive funding (€10k in Stage I and up to €250k in Stage II) to support early-stage ventures in creating the evidence and building the team needed to get next-level funding. Details and contacts on their website (https://www.incate.net/).
- These things aren’t sources of funds but would help you develop funding applications
- The Global AMR R&D Hub’s dynamic dashboard (link) summarizes the global clinical development pipeline, incentives for AMR R&D, and investors/investments in AMR R&D.
- Antimicrobial Resistance Research and Innovation in Australia is an actively updated summary that covers Australia’s AMR research and patent landscape. It is provided via collaboration between The Lens (an ambitious project seeking to discover, analyse, and map global innovation knowledge) and CSIRO (Commonwealth Scientific and Industrial Research Organisation, an Australian Government agency responsible for scientific research). Lots to explore here!
- Diagnostic developers would find valuable guidance in this 6-part series on in vitro diagnostic (IVD) development. Sponsored by CARB-X, C-CAMP, and FIND, it pulls together real-life insights into a succinct set of tutorials.
- In addition to the lists provided by the Global AMR R&D Hub, you might also be interested in my most current lists of R&D incentives (link) and priority pathogens (link).
John’s Top Recurring Meetings
Virtual meetings are easy to attend, but regular attendance at annual in-person events is the key to building your network and gaining deeper insight. My personal favorites for such in-person meetings are below. Of particular value for developers, the small meeting format of BEAM’s AMR Conference (March) and GAMRIC (September-October; formerly, the ESCMID-ASM conference series) creates excellent global networking. IDWeek (October) and ECCMID (April) are much larger meetings but also provide opportunities for networking with a substantial, focused audience via their Pipeline sessions. Hope to see you there!
- 1-3 Oct 2025 GAMRIC, the Global AMR Innovators Conference (London, UK). Formerly the ESCMID-ASM Joint Conference on Drug Development for AMR, this meeting series is now in its 10th year and is being continued under the joint sponsorship of CARB-X, ESCMID, BEAM Alliance, GARDP, LifeArc, Boston University, and AMR.Solutions. The ongoing series will continue the successful format of prior meetings with a single-track meeting and substantial networking time (go here to see details of the outstanding 2024 meeting).
- Registration is now open and the preliminary agenda can be found at that same link (https://www.gamric.org/). The meeting will be limited to approximately 300 attendees, so please be sure to register promptly to avoid disappointment!
- 19-22 Oct 2025 (Georgia, USA): IDWeek 2025, the annual meeting of the Infectious Diseases Society of America. Go here to register. For those who would like a substantial opportunity to present a product to a large audience (see also adjacent note about ESCMID), note the call for applications to present at an IDWeek Pipeline Session; go here to submit an application for your compound or diagnostic.
- 3-4 Mar 2026 (Basel, Switzerland): The 10th AMR Conference. Sponsored by the BEAM Alliance, the 9th AMR Conference has just concluded and it’s again been an excellent meeting! Please mark your calendar for next year. You can’t register yet, but details will appear here!
- 17-21 April 2026 (Munich, Germany): ESCMID Global 2026, the annual meeting of the European Society for Clinical Microbiology and Infectious Diseases. You can’t register yet, but you can go here for details on the outstanding 2025 meeting. For those who would like a substantial opportunity to present a product to a large audience (see also adjacent note about IDWeek), I know that the meeting schedule will again include Pipeline Monday; go here to see details from 2025.
Upcoming meetings of interest to the AMR community:
- 19 Aug 2022 (virtual, 11a-12.30p EST) Webinar entitled “Challenges and opportunities in the treatment of syphilis”, organized by GARDP REVIVE. Go here to register. Note also that recordings of prior REVIVE webinars are available for replay.
- 9 Sep 2022 (virtual, 9.30-11.00 CEST) Webinar entitled “Overcoming challenges of tuberculosis drug discovery and development”, organized by GARDP REVIVE. Go here to register. Note also that recordings of prior REVIVE webinars are available for replay.
- 10-11 Sep 2025 (Ghent, BE): Organized by a group at Ghent University in collaboration with the ESCMID Study Group for Non-Traditional Antibacterial Therapy (ESGNTA), the Phage Protein Meeting is an interdisciplinary conference dedicated to advancing the field of phage proteins for infection control that seeks to bring together academic researchers and professionals working with phage proteins such as phage lysins, tail fibers, tailspikes, depolymerases, and tailocins. Note that the focus here is on phage proteins rather than phage themselves. Go here for details and to register.
- 12 Sep 2025 (Ghent, BE): The 10-11 Sep 2025 meeting just above is followed in Ghent on 12 Sep by the 4th annual meeting of BSVoM (Belgian Society for Viruses of Microbes). This meeting is broader than therapeutics, providing an “interdisciplinary perspective on virus of microbes, ranging from basic research to industrial developments and clinical use.” Go here for details.
- 10-13 Sep 2025 (Lisbon, Portugal): 6th ESCMID Conference on Vaccines. Go here for details.
- 15-19 Sep 2025 (virtual): US CDC-sponsored the 9th Annual Fungal Disease Awareness Week (FDAW). Daily themed events include “Think Fungus” on 15 Sep, “Fungi are everywhere” on 16 Sep, and “Fungal diseases and drug resistance (19 Sep). Signup for the Mycotic Diseases Branch newsletter to stay updated.
- 1-3 Oct 2025 GAMRIC, the Global AMR Innovators Conference (London, UK; formerly the ESCMID-ASM Joint Conference on Drug Development for AMR). See list of Top Recurring meetings, above..
- 11-19 Oct 2025 (Annecy, France, residential in-person program): ICARe (Interdisciplinary Course on Antibiotics and Resistance) … and 2025 will be the 9th year for this program. Patrice Courvalin orchestrates content with the support of an all-star scientific committee and faculty. The resulting soup-to-nuts training covers all aspects of antimicrobials, is very intense, and routinely gets rave reviews! Seating is limited, so mark your calendars now if you are interested. Applications are being accepted from 20 Mar to 21 Jun 2025 — go here for more details.
- 17-20 Sep 2025 (Porto, PT): 14th International Meeting on Microbial Epidemiological Markers (IMMEM XIV). Go here for details.
- 9-13 Nov 2025 (Portland, OR, USA): ASM Conference on Biofilms. Go here for details and to register.
- 18-24 Nov 2025 (global, multiple locations): World Antibiotic Awareness Week (WAAW) is convened annually on 18-24 Nov by WHO with national events (e.g., CDC’s US Antibiotic Awareness Week (USAAW); ECDC’s 18 Nov European Antibiotic Awareness Day) occurring around the globe. Details will follow as events become visible.
- 19-22 Oct 2025 (Georgia, USA): IDWeek 2025. See list of Top Recurring meetings, above.
- 29-31 Oct 2025 (Bengalaru, India): ASM Global Research Symposium on the One Health Approach to Antimicrobial Resistance (AMR), hosted in partnership with the Centre for Infectious Disease Research (CIDR) at the Indian Institute of Science (IISc). Go here for details and to register.
- 28-30 Jan 2026 (Las Vegas, NV, USA): IDSA and ASM have announced a new US-based meeting series entitled IAMRI (Interdisciplinary Meeting on Antimicrobial Resistance and Innovation) and described as a “forum for collaboration and exploration around the latest advances in antimicrobial drug discovery and development.” You can’t register yet (the website says registration will open August 2025) but you go here for the program and to submit an abstract (deadline for abstracts is 1 Oct 2025).
- 3-4 Mar 2026 (Basel, Switzerland): The 10th AMR Conference sponsored by the BEAM Alliance. See list of Top Recurring meetings, above.
- 8-13 Mar 2026 (Renaissance Tuscany Il Ciocco, Italy): 2026 Gordon Research Conference (GRC) entitled “Antibacterials of Tomorrow to Combat the Global Threat of Antimicrobial Resistance.” A Gordon Research Seminar (GRS) will be held the weekend before (7-8 Mar) for young doctoral and post-doctoral researchers. Space for the GRS and the GRC is limited; for details and to apply, go here for the GRC and here for the GRS.
- 17-21 April 2026 (Munich, Germany): ESCMID Global 2026, the annual meeting of the European Society for Clinical Microbiology and Infectious Diseases. See Recurring Meetings list, above.
- 4-8 June 2026 (Washington, DC): ASM Microbe, the annual meeting of the American Society for Microbiology. The meeting format is evolving and next year will combine 3 meetings (ASM Health, ASM Applied and Environmental Microbiology, and ASM Mechanism Discovery) into one event. Go here for details.
Self-paced courses, online training materials, and other reference materials:
- OpenWHO: “Antimicrobial Resistance in the environment: key concepts and interventions.” Per the webpage for the course, it will teach you “…why addressing AMR in the environment is essential and gain insights into how action can be taken to prevent and control AMR in the environment at the national level.” This course builds on WHO’s 2024 Guidance on wastewater and solid waste management for manufacturing of antibiotics. For further reading, see also the 25 Sep 2023 newsletter entitled “Manufacturing underpins both access and stewardship: Cefiderocol as a case study” and the 28 Jan 2024 newsletter entitled “EMA Concept Paper: Guidance on manufacturing of phage products”.
- GARDP’s REVIVE website provides an encyclopedia covering a range of R&D terms, recordings of prior GARDP webinars, a variety of viewpoint articles, and more! Check it out!
- GARDP’s https://antibioticdb.com/ is an open-access database of antibacterial agents.
- The CARB-X website provides a range of recordings from its webinars, bootcamps, and more. A bit of browsing would be time well spent!
- British Society for Antimicrobial Chemotherapy offers an eLearning section: Education – The British Society for Antimicrobial Chemotherapy.